CTA Hypoperfusion Analysis
Upload your CT angiography brain to automatically compute the hypoperfused area relevant from stroke care
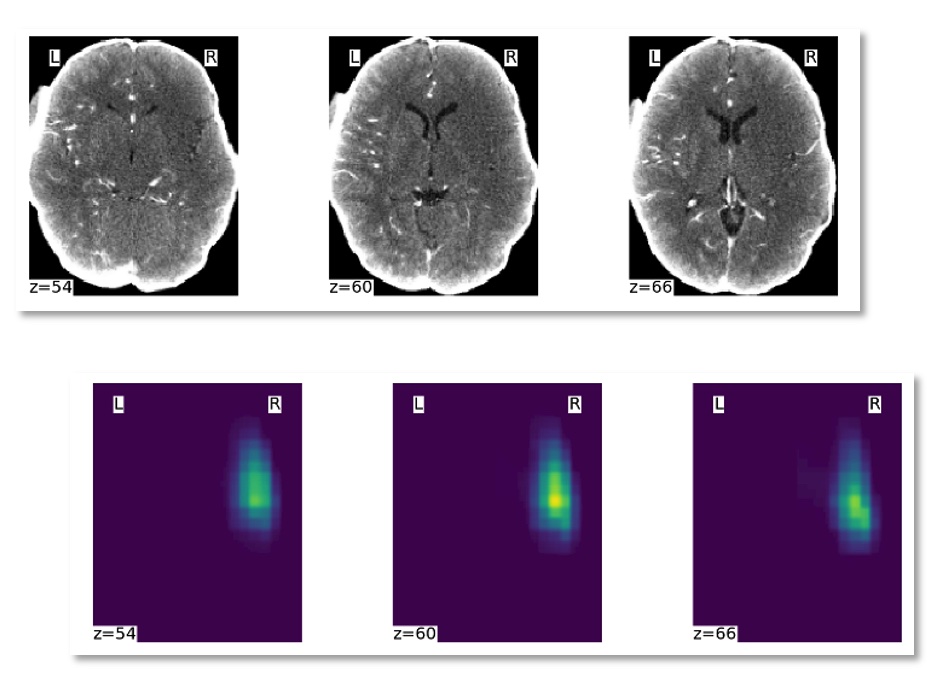
Upload your CT angiography brain to automatically compute the hypoperfused area relevant from stroke care

This software requires CTA brain images in compressed NIFTI format (i.e. with a nii.gz extension) that have been pre-registered to
a common brain template. The brain templates are available here
and here.
We have developed a point and click pipeline to perform the NIFTI format conversion, automatic registration and anonymization. It is available at
https://github.com/lgiancaUTH/preproc__cta_core .
A video showing how to use the pipeline works is available here.
If you would like to quickly test the software, you can download two example CTAs that have been already registered to the template:
here and
here .
A video showing how to use this Web App is available here.
Select your CTA image and upload it using the button below. The images need to be registered to a custom template, click here for more information. You can test our system using an example image. You can upload up to 5 images.
If you use this tool for your work, please do cite our publication: Giancardo L,
Niktabe A, Ocasio L, Abdelkhaleq R, Salazar-Marioni S, Sheth SA. Segmentation of acute stroke infarct core using image-level labels on CT-angiography.
NeuroImage: Clinical 2023;37:103362. https://doi.org/10.1016/j.nicl.2023.103362 .
This work is supported by the NIH NINDS R01NS121154. It has been developed by the Giancardo Lab in collaboration with the Center for Secure Artificial intelligence For hEalthcare (SAFE) and Sheth Lab. Special thanks to UTHealth the Institute for Stroke and Cerebrovascular Diseases.